Creating CellTypeDatasets
Alan Murphy, Brian Schilder, and Nathan Skene
Oct-22-2021
Source: vignettes/create_CTD.Rmd
create_CTD.RmdIntroduction
EWCE was originally designed to work with the single-cell cortical transcriptome data from the Linnarsson Lab1 (available here).
Using this package it is possible to read in any single-cell transcriptome data, provided that you have a cell by gene expression matrix (with each cell as a separate column) and a separate annotation dataframe, with a row for each cell.
The EWCE process involves testing for whether the genes in a target list have higher levels of expression in a given cell type than can reasonably be expected by chance. The probability distribution for this is estimated by randomly generating gene lists of equal length from a set of background genes.
EWCE can be applied to any gene list. In the original paper we reported its application to genetic and transcriptomic datasets..2
Note that throughout this vignette we use the terms ‘cell type’ and ‘cell subtype’ to refer to two levels of annotation of what a cell type is. This is described in further detail in our paper2, but relates to the two levels of annotation provided in the Linnarsson dataset.1 In this dataset a cell is described as having a cell type (i.e. ‘Interneuron’) and subcell type (i.e. ‘Int11’ a.k.a Interneuron type 11).
Creating a CellTypeDataset
Here we provide an example of converting a scRNA-seq dataset (Zeisel 2015) into a CellTypeDataset (CTD) so it can used with EWCE.
Loading datasets
The first step for all analyses is to load the single-cell transcriptome (SCT) data. For the purposes of this example we will use the dataset described in: > Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq, Science, 2015
This is stored and can be loaded as follows from ewceData package.
EWCE can generate CTD from two different data input types: 1. A gene x cell expression matrix (can be dense matrix, sparse matrix, data.frame, or DelayedArray).
2. A SingleCellExperiment (SCE) or SummarizedExperiment object.
To check the data, we can quickly plot the distribution of expression of a given gene across all the cell types.
# example datasets held in ewceData package
cortex_mrna <- ewceData::cortex_mrna()
gene <- "Necab1"
cellExpDist <- data.frame(Expression=cortex_mrna$exp[gene,],
Celltype=cortex_mrna$annot[
colnames(cortex_mrna$exp),]$level1class)
ggplot(cellExpDist) +
geom_boxplot(aes(x=Celltype, y=Expression), outlier.alpha = .5) +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
theme_bw()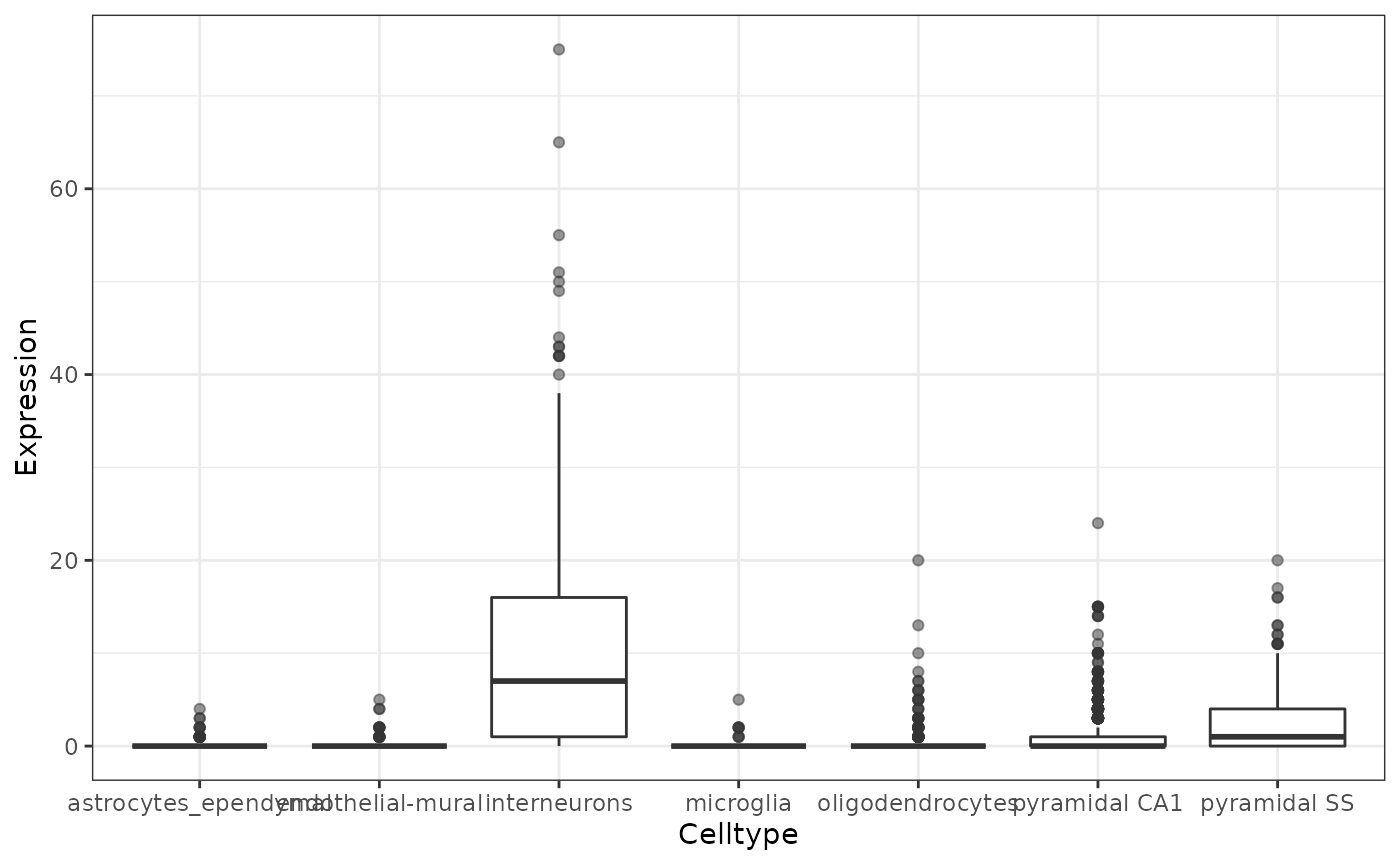
To note for the analysis of the cortex_mrna dataset, I will proved the alternative for an SCE object under each segment of code if there is any difference.
Converting single-cell formats
If you would like to try running the functions on an SCE object instead, we can convert cortex_mrna with the package scKirby. scKirby automatically infers the input file type and converts it to SCE format by default.
if (!"SingleCellExperiment" %in% rownames(installed.packages())){BiocManager::install("SingleCellExperiment")}
library(SingleCellExperiment)
if (!"scKirby" %in% rownames(installed.packages())){remotes::install_github("bschilder/scKirby")}
library(scKirby)
# Make SCE object
cortex_mrna <- ewceData::cortex_mrna()
cortex_mrna_sce <- scKirby::ingest_data(cortex_mrna, save_output = FALSE)
# Now to plot the SCE dataset:
gene="Necab1"
cellExpDist_sce = data.frame(e=assays(cortex_mrna_sce)[[1]][gene,],
l1=colData(cortex_mrna_sce)[
colnames(assays(cortex_mrna_sce)[[1]]),
]$level1class)
ggplot(cellExpDist_sce) + geom_boxplot(aes(x=l1,y=e)) + xlab("Cell type") +
ylab("Unique Molecule Count") +
theme(axis.text.x = element_text(angle = 90, hjust = 1)) Correcting gene symbols
It is very common for publicly available transcriptome datasets to use incorrect gene symbols (often some gene names will have been mangled by opening in Excel).
A function is provided to correct these where out of date aliases were used and give a warning if appears possible that excel has mangled some of the gene names. This function can be used with a downloaded reference file like the MRK_List2 file from MGI which lists known synonyms for MGI gene symbols. If no file path is passed, there is a loaded reference dataset of the MRK_List2 which will be used: mgi_synonym_data(). We recommend running this function on all input datasets.
cortex_mrna$exp = fix_bad_mgi_symbols(cortex_mrna$exp)For the SCE version, pass the whole SCE object
#Note that data in SCE object must be in 'counts' assay
#The fix_bad_hgnc_symbols function can similarly take an SCE object as an input
cortex_mrna_sce = fix_bad_mgi_symbols(cortex_mrna_sce)Drop genes
The vignette takes a while to go through if you use all genes. So let’s restrict to 1000 random genes.
nKeep <- 1000
must_keep <- c("Apoe","Gfap","Gapdh")
keep_genes <- c(must_keep,sample(rownames(cortex_mrna$exp),997))
cortex_mrna$exp <- cortex_mrna$exp[keep_genes,]
dim(cortex_mrna$exp)## [1] 1000 3005#SCE
nKeep = 1000
must_keep = c("Apoe","Gfap","Gapdh")
keep_genes = c(must_keep,sample(names(rowRanges(cortex_mrna_sce)),997))
cortex_mrna_sce = cortex_mrna_sce[keep_genes,]Normalization
A different number of reads is found across each cell. We suggest using scTransform to normalise for differences due to cell size, then linearly scale. Note that this might be slow.
cortex_mrna$exp_scT_normed <- EWCE::sct_normalize(cortex_mrna$exp) ##
|
| | 0%## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached##
|
|=================================== | 50%## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached
## Warning in theta.ml(y = y, mu = fit$fitted): iteration limit reached##
|
|======================================================================| 100%
##
|
| | 0%
|
|=================================== | 50%
|
|======================================================================| 100%
##
|
| | 0%
|
|=================================== | 50%
|
|======================================================================| 100%
# SCE
SummarizedExperiment::assays(cortex_mrna_sce)$normcounts <- EWCE::sct_normalize(SummarizedExperiment::assays(cortex_mrna_sce)$counts)Note that this is optional (and was not used in the original EWCE publication) so by all means ignore this scTransform step.
Calculate specificity matrices
The next steps are as follows:
-
drop_uninformative_genes(): Drop genes which do not show significant evidence of varying between level 2 cell types (based on ANOVA). Optionally, you can also drop all genes which do not have 1:1 mouse:human orthologues (Note: this used to a separate step but has since been integrated directly intodrop_uninformative_genes()via the argumentsdrop_nonhuman_genes=andinput_species=). - Calculate cell type averages and specificity for each gene.
The last step is only necessary if you plan to compare to human data, i.e. gene sets resulting from human genetics.
Rather than returning the data directly, the functions save the calculated data to a file and the file name is returned.
# Generate cell type data for just the cortex/hippocampus data
exp_CortexOnly_DROPPED <- EWCE::drop_uninformative_genes(
exp = cortex_mrna$exp_scT_normed,
input_species = "mouse",
output_species = "human",
level2annot = cortex_mrna$annot$level2class) ## Time difference of 0.2545247 secsGenerate CellTypeDataset
annotLevels <- list(level1class=cortex_mrna$annot$level1class,
level2class=cortex_mrna$annot$level2class)
fNames_CortexOnly <- EWCE::generate_celltype_data(
exp = exp_CortexOnly_DROPPED,
annotLevels = annotLevels,
groupName = "kiCortexOnly",
savePath = tempdir())
ctd_CortexOnly <- EWCE::load_rdata(fNames_CortexOnly)#SCE
# Generate cell type data for just the cortex/hippocampus data
exp_CortexOnly_DROPPED <- drop_uninformative_genes(exp=cortex_mrna_sce,
drop_nonhuman_genes = T,
input_species = "mouse",
level2annot=cortex_mrna_sce$level2class)
annotLevels = list(level1class=cortex_mrna_sce$level1class,
level2class=cortex_mrna_sce$level2class)
fNames_CortexOnly <- generate_celltype_data(exp=exp_CortexOnly_DROPPED,
annotLevels=annotLevels,
groupName="kiCortexOnly",
savePath=tempdir()) To note on the SCE approach, these are the only differences for handling the analysis of an SCE object. All downstream analysis on cortex_mrna would be the same. The same approach can be used for other datasets in the vignette if converted such as hypothalamus_mrna or your own SCE object.
Merging two single-cell datasets
Often it is useful to merge two single-cell datasets. For instance, there are separate files for the cortex and hypothalamus datasets generated by the Karolinska institute. A subset of the resulting merged dataset is loaded using ctd() but it is worth understanding the approach (outlined below) if you want to repeat this for other single-cell datasets.
The dataset is available from the GEO but is also available in the ewceData package: hypothalamus_mrna(). See the ewceData for details of how this file was preprocessed after first being downloaded from GEO, unzipped and read into R.
We can now merge the hypothalamus dataset with the cortex dataset and then calculate specificity:
Load hypothalamus dataset
hypothalamus_mrna <- ewceData::hypothalamus_mrna()Fix bad MGI symbols
hypothalamus_mrna$exp <- EWCE::fix_bad_mgi_symbols(exp = hypothalamus_mrna$exp)
cortex_mrna$exp <- EWCE::fix_bad_mgi_symbols(exp = cortex_mrna$exp)Merge CTD
Merge the CTDs - again this can be run with SCE or other ranged SE objects as input.
merged_KI <- EWCE::merge_two_expfiles(exp1 = hypothalamus_mrna$exp,
exp2 = cortex_mrna$exp,
annot1 = hypothalamus_mrna$annot,
annot2 = cortex_mrna$annot,
name1 = "Hypothalamus (KI)",
name2 = "Cortex/Hippo (KI)")Drop uninformative genes
Drop genes which don’t vary significantly between cell types.
exp_merged_DROPPED <- EWCE::drop_uninformative_genes(
exp = merged_KI$exp,
drop_nonhuman_genes = TRUE,
input_species = "mouse",
level2annot = merged_KI$annot$level2class)## Time difference of 0.6415126 secsGenerate CellTypeDataset
annotLevels = list(level1class=merged_KI$annot$level1class,
level2class=merged_KI$annot$level2class)
# Update file path to where you want the cell type data files to save on disk
fNames_MergedKI <- EWCE::generate_celltype_data(exp = exp_merged_DROPPED,
annotLevels = annotLevels,
groupName = "MergedKI",
savePath = tempdir())
ctd_MergedKI <- load_rdata(fNames_MergedKI) Understanding specificity matrices
While not required for further analyses it helps to understand what the outputs of this function are.
Note firstly, that it is a list such that ctd[[1]] contains data relating to level 1 annotations and ctd[[2]] relates to level 2 annotations.
Using the ggplot2 package to visualise the data, let us examine the expression of a few genes. If you have not already done so you will need to first install the ggplot2 package with install.packages("ggplot2").
For this example we use a subset of the genes from the merged dataset generated above, which is accessed using ewceData::ctd(). We recommend that you use the code above to regenerate this though and drop the first command from the below section.
## Load merged cortex and hypothalamus dataset generated by Karolinska institute
ctd <- ewceData::ctd() # i.e. ctd_MergedKI
plt <- EWCE::plot_ctd(ctd = ctd,
level = 1,
genes = c("Apoe","Gfap","Gapdh"),
metric = "mean_exp")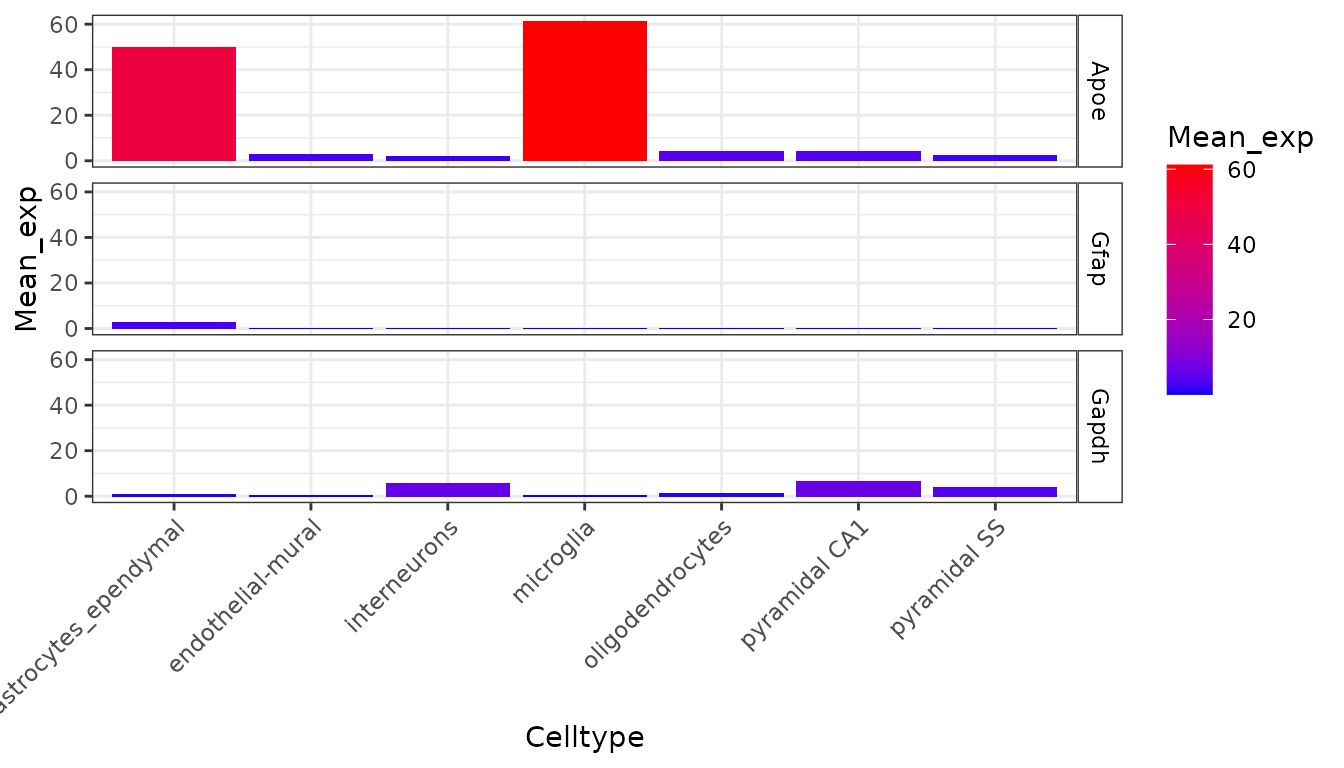
This graph shows the average expression of three genes: Apoe, Gfap and Gapdh. While there are substantial differences in which cell types express these genes, the dominant effect seen here is the overall expression level of the data. For the purposes of this analysis though, we are not interested in overall expression level and only wish to know about the proportion of a genes expression which is found in a particular cell type. We can study this instead using the following code which examines the data frame ctd[[1]]$specificity:
plt <- EWCE::plot_ctd(ctd = ctd,
level = 1,
genes = c("Apoe","Gfap","Gapdh"),
metric = "specificity")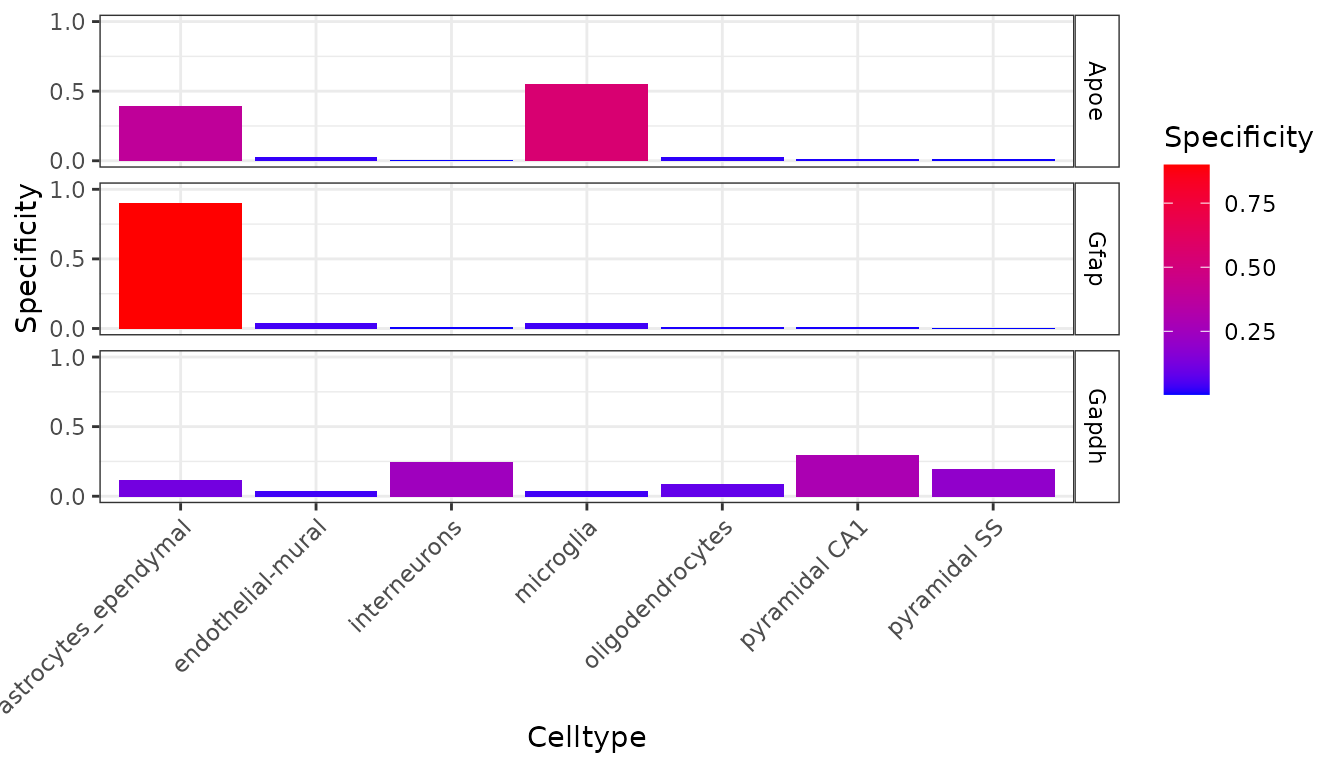
We can now see in this graph that Gfap is the most specific to a cell type (Type 1 Astrocytes) of any of those three genes, with over 60% of its expression found in that cell type.
It can also be seen that the majority of expression of Gapdh is in neurons but because their are a greater number of neuronal subtypes, the total expression proportion appears lower. We can examine expression across level 2 cell type level annotations by looking at ctd[[2]]$specificity:
plt <- EWCE::plot_ctd(ctd = ctd,
level = 2,
genes = c("Apoe","Gfap","Gapdh"),
metric = "specificity")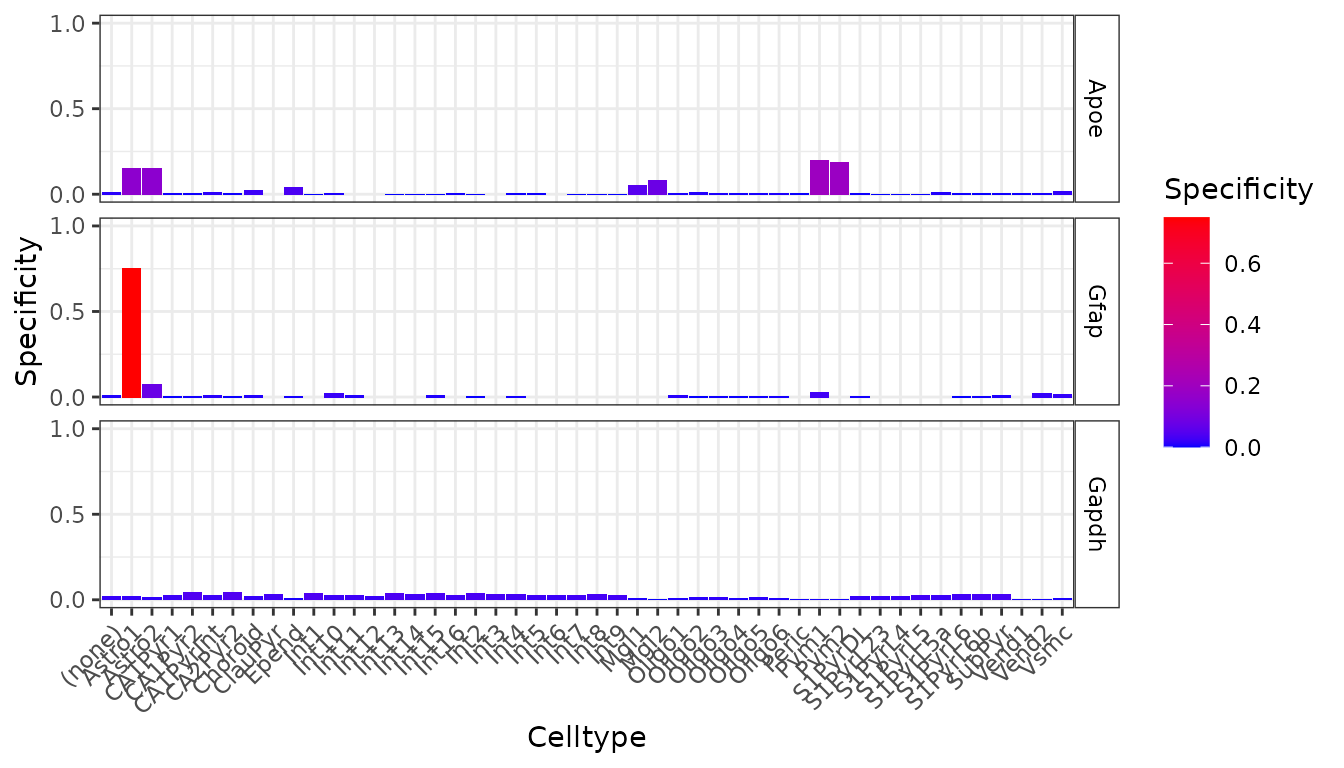
Session Info
utils::sessionInfo()## R version 4.1.1 (2021-08-10)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 20.04.2 LTS
##
## Matrix products: default
## BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.8.so
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=C
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] ewceData_1.1.0 ExperimentHub_2.1.4 AnnotationHub_3.1.6
## [4] BiocFileCache_2.1.1 dbplyr_2.1.1 BiocGenerics_0.39.2
## [7] ggplot2_3.3.5 EWCE_2.0.0 RNOmni_1.0.0
## [10] BiocStyle_2.21.4
##
## loaded via a namespace (and not attached):
## [1] readxl_1.3.1 backports_1.2.1
## [3] systemfonts_1.0.3 plyr_1.8.6
## [5] lazyeval_0.2.2 orthogene_0.99.8
## [7] BiocParallel_1.27.17 listenv_0.8.0
## [9] GenomeInfoDb_1.29.10 digest_0.6.28
## [11] htmltools_0.5.2 fansi_0.5.0
## [13] magrittr_2.0.1 memoise_2.0.0
## [15] openxlsx_4.2.4 limma_3.49.4
## [17] globals_0.14.0 Biostrings_2.61.2
## [19] matrixStats_0.61.0 pkgdown_1.6.1
## [21] colorspace_2.0-2 blob_1.2.2
## [23] rappdirs_0.3.3 textshaping_0.3.6
## [25] haven_2.4.3 xfun_0.27
## [27] dplyr_1.0.7 crayon_1.4.1
## [29] RCurl_1.98-1.5 jsonlite_1.7.2
## [31] glue_1.4.2 gtable_0.3.0
## [33] zlibbioc_1.39.0 XVector_0.33.0
## [35] HGNChelper_0.8.1 DelayedArray_0.19.4
## [37] car_3.0-11 future.apply_1.8.1
## [39] SingleCellExperiment_1.15.2 abind_1.4-5
## [41] scales_1.1.1 DBI_1.1.1
## [43] rstatix_0.7.0 Rcpp_1.0.7
## [45] viridisLite_0.4.0 xtable_1.8-4
## [47] foreign_0.8-81 bit_4.0.4
## [49] stats4_4.1.1 htmlwidgets_1.5.4
## [51] httr_1.4.2 ellipsis_0.3.2
## [53] farver_2.1.0 pkgconfig_2.0.3
## [55] sass_0.4.0 utf8_1.2.2
## [57] labeling_0.4.2 tidyselect_1.1.1
## [59] rlang_0.4.12 reshape2_1.4.4
## [61] later_1.3.0 AnnotationDbi_1.55.2
## [63] munsell_0.5.0 BiocVersion_3.14.0
## [65] cellranger_1.1.0 tools_4.1.1
## [67] cachem_1.0.6 generics_0.1.0
## [69] RSQLite_2.2.8 broom_0.7.9
## [71] ggdendro_0.1.22 evaluate_0.14
## [73] stringr_1.4.0 fastmap_1.1.0
## [75] yaml_2.2.1 ragg_1.1.3
## [77] babelgene_21.4 knitr_1.36
## [79] bit64_4.0.5 fs_1.5.0
## [81] zip_2.2.0 purrr_0.3.4
## [83] KEGGREST_1.33.0 gprofiler2_0.2.1
## [85] future_1.22.1 mime_0.12
## [87] compiler_4.1.1 plotly_4.10.0
## [89] filelock_1.0.2 curl_4.3.2
## [91] png_0.1-7 interactiveDisplayBase_1.31.2
## [93] ggsignif_0.6.3 tibble_3.1.5
## [95] bslib_0.3.1 homologene_1.4.68.19.3.27
## [97] stringi_1.7.5 highr_0.9
## [99] desc_1.4.0 forcats_0.5.1
## [101] lattice_0.20-45 Matrix_1.3-4
## [103] vctrs_0.3.8 pillar_1.6.4
## [105] lifecycle_1.0.1 BiocManager_1.30.16
## [107] jquerylib_0.1.4 data.table_1.14.2
## [109] bitops_1.0-7 httpuv_1.6.3
## [111] patchwork_1.1.1 GenomicRanges_1.45.0
## [113] R6_2.5.1 bookdown_0.24
## [115] promises_1.2.0.1 gridExtra_2.3
## [117] rio_0.5.27 IRanges_2.27.2
## [119] parallelly_1.28.1 codetools_0.2-18
## [121] MASS_7.3-54 assertthat_0.2.1
## [123] SummarizedExperiment_1.23.5 rprojroot_2.0.2
## [125] withr_2.4.2 sctransform_0.3.2
## [127] S4Vectors_0.31.5 GenomeInfoDbData_1.2.7
## [129] parallel_4.1.1 hms_1.1.1
## [131] grid_4.1.1 tidyr_1.1.4
## [133] rmarkdown_2.11 MatrixGenerics_1.5.4
## [135] carData_3.0-4 ggpubr_0.4.0
## [137] Biobase_2.53.0 shiny_1.7.1